Table of Contents
Keywords
•
Drug repositioning
•
Drugging undruggable targets
•
In silico candidate selection with in vitro, in vivo validation
•
TCGA lung cancer patient data
•
Project Achilles data
•
CMap data
Introduction
An in silico approach for the use of undruggable targets in cancer therapies and for revealing the underlying mechanisms of these targets.
In silico approach based on large-scale gene expression signatures to identify drug candidates for lung cancer metastasis.
GALNT14 was identified as a putative driver of lung cancer metastasis, leading to poor survival.
To overcome the poor druggability of GALNT14, they utilized Connectivity Map and identified bortezomib(BTZ) as a potent metastatic inhibitor, to target the downstream transcription factor of GALNT, bypassing the direct inhibition.
The antimetastatic effect of BTZ was verified both in vitro and in vivo.
Both BTZ treatment and GALNT14 knockdown attenuated TGF-mediated gene expression and suppressed TGF-dependent metastatic genes.
CMap: a collection of genome-wide expression profiles of cell lines treated with >20,000 chemicals
N-acetyl-galactosaminyltransferases(GALNTs) are key enzymes that initiate O-linked N-acetyl galactosamine glycosylation. This is an important step in the synthesis of Thomsen nouvelle(Tn) antigens, which are well-characterized tumor associated molecules. GALNT14 expression is a prognostic marker in neuroblastoma and lung cancer, and also a predictive marker for Apo2L/TRAIL-based cancer therapy.
Apo2L/TRAIL: a death receptor ligand(Apo2 ligand/TNF(tumor necrosis factor) related apoptosis-inducing ligand). This approach offers promising therapeutic potential based on its ability to induce apoptosis in various cancer cell lines with little toxicity toward normal cells.
Results
GALNT14 as a putative molecular target for lung cancer metastasis
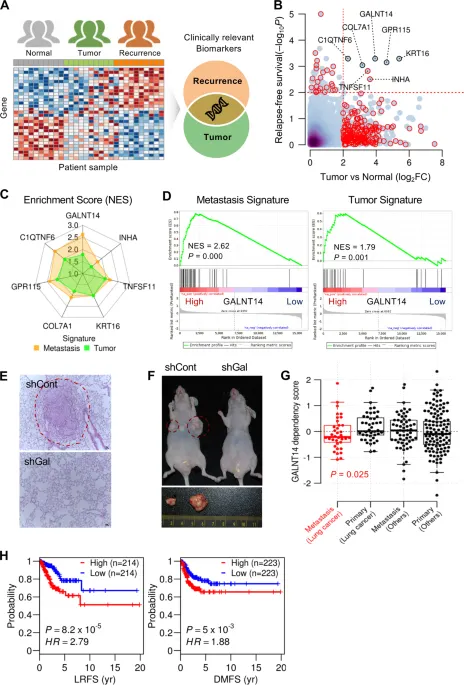
Assembled transcriptome data and clinical information from 516 lung cancer patients from the TCGA LUAD cohort. Fig 1a
Performed series of relapse-free survival(RFS) and differential expression analyses. Fig 1b
Expression of seven genes(GALNT14, COL7A1, GPR115, C1QTNF6, KRT16, INHA, TNFSF11) were significantly associated with cancer progression ,recurrence, and overall survival.
Divided lung cancer patients into two groups(low / high expression of those genes).
Both metastasis and tumor signatures were positively enriched in high-expression group. Fig 1c
Significant enrichment was also observed for GALNT14. Fig 1d
Several other studies reveal that GALNT14 contributes to both tumor malignancy and metastasis. Fig 1e(metastatic), Fig 1f(tumorigenic potential)
Metastatic lung cancer cells were more vulnerable to GALNT14 depletion than non-metastatic or other types of cancer in the Project Achilles dataset. Fig 1g
GALNT14 expression showed clear negative correlation with both locoreginal recurrence-free survival(LRFS), and distant metastasis-free survival(DMFS). Fig 1h
These results suggest that GALNT14 is a promising molecular target for lung cancer metastasis that can be utilized to improve patient survival.
Computational repositioning of BTZ to reverse the GALNT14 expression signature
There have been a few attemps to identify specific inhibitors for GALNT14, but failed due to poor druggability.
Notably, GALNT14-dependent metastatic potential is governed by the induction of transcription factors(HOXB9, SOX4).
HOXB9 and SOX4 are transcription factor genes regulated by GALNT14.
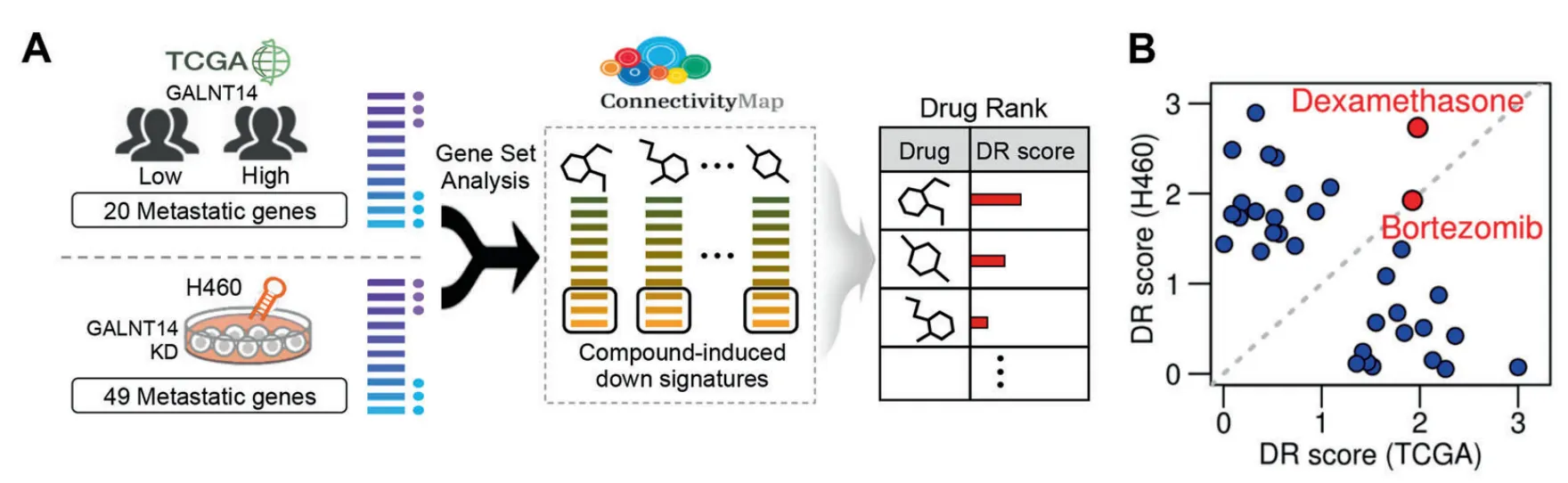
Thus, rather than directly inhibiting the enzymatic activity of GALNT14, they utilized CMap dataset to virtually screen drugs that mimic the effects of GALNT14 depletion at the transcriptome level.
Among 3711 metastasis-related genes, they selected two gene sets
1) 20 genes upregulated in the GALNT14-high group in TCGA
2) 49 genes downregulated by GALNT14 knockdown in the H460 cell ine
They performed two independent predictions using CMap analysis. Candidate drugs were prioritized according to their DR scores. Fig 2a
Dexamethasone(DEX) and BTZ were among the top candidates in both predictions. Fig 2b
Of the differentially expressed genes in response to either DEX or BTZ in the CMap dataset, SOX4 was altered by both, which suggest that SOX4 could serve as a validation marker.
They performed cell migration test for two drugs, and observed that BTZ treatment led to a significant reduction in migration capacity of H460 cells, whereas DEX did not.
This suggest that initial screening with marker gene is important for further in vitro analysis.
for proteasome inhibition: 22 nM - Fig 2d
for migration inihibition: 17 nM - Fig 2e
Observed invasion inhibition at 20 nM - Fig 2f
At 20 nM of BTZ, there were no significant change in cell viability or cell proliferation.
They also found out that a cell line that has lower GALNT14 expression did not respond to BTZ treatment in terms of migration or invasion. Which suggest that anti-invasion/migration effect of BTZ is associated with GALNT14 expression.
The effect of BTZ in relation to proteasome inhibition
Since anti-invasion/migration effect occurred at a concentration that also inhibited proteasome, they wanted to determine wheter this effect was the direct result of proteasome inhibition.
To investigate this, they selected three FDA-approved proteasome inhibitors, BTZ, Carfilzomib(CFZ), Ixazomib(IXZ), all approved for the treatment of multiple myeloma.
CFZ inhibited proteasome activity, but not migration or invasion.
IXZ inhibited proteasome activity, but not migration.
These data suggest that the observed anti-migration/invation effect of BTZ may arise from its off-target effect and that is independent of proteasome inhibition.
To confirm this, they also compared the transcriptome profiles of H460 cells treated with BTZ or CFZ, or depleted of GALNT14(shGAL, knockdown of GALNT14). - Fig 3e
Perturbation by BTZ and shGAL(but not CFZ) induced similar transcriptomic changes relative to the control.
The expression patterns of the metastasis-related genes were even more similar between BTZ and shGal.
In contrast, proteasome related genes were altered significantly by both drugs, but only marginally for shGAL.
The genes downregulated by BTZ and shGAL overlapped significantly, suggesting that BTZ treatment partially mimics the depletion of GALNT14. - Fig 3f
Attenuation of the TGF gene response by BTZ treatment or GALNT14 knockdown
Hypothesis: a subset of the 101 genes down regulated by both BTZ treatment and GALNT14 depletion could account for the anti-migration/invasion effects of BTZ
They conducted pathway enrichment analysis of the 101 genes and investigated the clinical significance of each pathway. - Fig 4a
Most enriched pathway was TGF signaling. They performed various experiments and found out that the suppression of TGF signaling and gene expression responses by BTZ makes a major contribution to reduced migration and invasion.
In vivo validation of the antimetastatic effect of BTZ
Given the anti-migration/invasion effect of BTZ in a lung cancer cell model, they tested the in vivo efficacy of BTZ against cancer metastasis.
For this purpose, local metastasis was induced in mice via the tail vein injection of H460 lung cancer cells, followed by treatment with or without BTZ or CFZ. - Fig 5a
The proteasome-inhibitory effect of BTZ and CFZ was examined by measuring proteasome activity in blood. - Fig 5b
BTZ treatment was found to considerably attenuate lung colonization(Fig 5g) with a proteasome inhibitory effect(Fig 5h) but no physiological abnormalities.
Discussion
Unlike other studies that used CMap, they focused exclusively on a target gene related to a pertinent phenotype and identified BTZ as a drug candidate with novel anti-metastatic effects.
Previous studies used CMap method to analyze differences in gene expression signatures between normal and cancerous tissue.
They demonstrated that anti-metastatic effect of BTZ is distinct from its proteasome inhibition.
In pathway level, the most enriched pathway was TGF signaling, and they also identified the GALNT14-TGF signature, which has invasive properties that are attenuated by BTZ.
Accurate determination of gene signature that represents the target phenotype based on molecular mechanisms is an important prerequisite step to minimize the number of false positives.
This could be conducted using in silico DR tools.
They integrated multiple independent expression signatures from cancer patients(TCGA), genetic perturbations(knock-down or overexpression), and drug treatment(CMap). This strategy is generally applicable to other types of target. This is the first proof-of-concept for this DR approach.